SARS-related coronavirus
Template:Short description Template:About Template:Use Oxford spelling Template:Use dmy dates Template:Virusbox
Severe acute respiratory syndrome–related coronavirus (SARSr-CoV or SARS-CoV, Betacoronavirus pandemicum)<ref>Template:Cite web</ref><ref group=note>The terms SARSr-CoV and SARS-CoV are sometimes used interchangeably, especially prior to the discovery of SARS-CoV-2. This may cause confusion when some publications refer to SARS-CoV-1 as SARS-CoV.</ref> is a species of virus consisting of many known strains. Two strains of the virus have caused outbreaks of severe respiratory diseases in humans: severe acute respiratory syndrome coronavirus 1 (SARS-CoV or SARS-CoV-1), the cause of the 2002–2004 outbreak of severe acute respiratory syndrome (SARS), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of the pandemic of COVID-19.<ref name="CVSG">Template:Cite journal</ref><ref name="SCI">Template:Cite journal</ref> There are hundreds of other strains of SARSr-CoV, which are only known to infect non-human mammal species: bats are a major reservoir of many strains of SARSr-CoV; several strains have been identified in Himalayan palm civets, which were likely ancestors of SARS-CoV-1.<ref name="CVSG" /><ref>Template:Cite journal</ref><ref>Template:Cite news</ref><ref>Template:Cite journal</ref>
These enveloped, positive-sense single-stranded RNA viruses enter host cells by binding to the angiotensin-converting enzyme 2 (ACE2) receptor.<ref name="pmid24172901"/> The SARSr-CoV species is a member of the genus Betacoronavirus and the only species of the subgenus Sarbecovirus (SARS Betacoronavirus).<ref name="ICTV10">Template:Cite web</ref><ref name=":02">Template:Cite journal</ref>
The SARS-related coronavirus was one of several viruses identified by the World Health Organization (WHO) in 2016 as a likely cause of a future epidemic in a new plan developed after the Ebola epidemic for urgent research and development before and during an epidemic towards diagnostic tests, vaccines and medicines. This prediction came to pass with the COVID-19 pandemic.<ref>Template:Cite web</ref><ref>Template:Cite web</ref>
Classification
SARS-related coronavirus is a member of the genus Betacoronavirus (group 2) and monotypic of the subgenus Sarbecovirus (subgroup B).<ref>Template:Cite journal</ref> Sarbecoviruses, unlike embecoviruses or alphacoronaviruses, have only one papain-like proteinase (PLpro) instead of two in the open reading frame ORF1ab.<ref>Template:Cite journal</ref> SARSr-CoV was determined to be an early split-off from the betacoronaviruses based on a set of conserved domains that it shares with the group.<ref>Template:Cite journal</ref><ref>Template:Cite web</ref>
Bats serve as the main host reservoir species for the SARS-related coronaviruses like SARS-CoV-1 and SARS-CoV-2. The virus has coevolved in the bat host reservoir over a long period of time.<ref>Template:Cite journal</ref> Only recently have strains of SARS-related coronavirus been observed to have evolved into having been able to make the cross-species jump from bats to humans, as in the case of the strains SARS-CoV-1 and SARS-CoV-2.<ref name="pmid18258002">Template:Cite journal</ref><ref name="pmid24172901">Template:Cite journal</ref> Both of these strains descended from a single ancestor but made the cross-species jump into humans separately. SARS-CoV-2 is not a direct descendant of SARS-CoV-1.<ref name="CVSG" />
Genome

The SARS-related coronavirus is an enveloped, positive-sense, single-stranded RNA virus. Its genome is about 30 kb, which is one of the largest among RNA viruses. The virus has 14 open reading frames which overlap in some cases.<ref name="Snijder_2003">Template:Cite journal</ref> The genome has the usual 5′ methylated cap and a 3′ polyadenylated tail.<ref name = "Fehr_2015">Template:Cite book</ref> There are 265 nucleotides in the 5'UTR and 342 nucleotides in the 3'UTR.<ref name="Snijder_2003" />
The 5' methylated cap and 3' polyadenylated tail allows the positive-sense RNA genome to be directly translated by the host cell's ribosome on viral entry.<ref>Template:Cite book</ref> SARSr-CoV is similar to other coronaviruses in that its genome expression starts with translation by the host cell's ribosomes of its initial two large overlapping open reading frames (ORFs), 1a and 1b, both of which produce polyproteins.<ref name="Snijder_2003" />
| Function of SARSr-CoV genome proteins | |
|---|---|
| Protein | Function<ref name=McBride2012_Table1>Template:Cite journal</ref><ref name="Tang_2009">Template:Cite journal</ref><ref>Template:Cite journal</ref><ref name="redondo_2021">Template:Cite journal</ref> |
| ORF1ab Template:UniProt |
Replicase/transcriptase polyprotein (pp1ab) (nonstructural proteins) |
| ORF2 Template:UniProt |
Spike (S) protein, virus binding and entry (structural protein) |
| ORF3a Template:UniProt |
Interacts with S, E, M structural proteins; Ion channel activity; Upregulates cytokines and chemokines such as IL-8 and RANTES; Upregulates NF-κB and JNK; Induces apoptosis and cell cycle arrest, via Caspase 8 and -9, and by Bax, p53, and p38 MAP kinase |
| ORF3b Template:UniProt |
Upregulates cytokines and chemokines by RUNX1b; Inhibits Type I IFN production and signaling; Induces apoptosis and cell cycle arrest; |
| ORF3c Template:UniProt |
Unknown; first identified in SARS-CoV-2 but also present in SARS-CoV |
| ORF3d Template:UniProt |
Novel gene in SARS-CoV-2, of unknown function |
| ORF4 Template:UniProt |
Envelope (E) protein, virus assembly and budding (structural protein) |
| ORF5 Template:UniProt |
Membrane (M) protein, virus assembly and budding (structural protein) |
| ORF6 Template:UniProt |
Enhances cellular DNA synthesis; Inhibits Type I IFN production and signaling |
| ORF7a Template:UniProt |
Inhibits cellular protein synthesis; Induces inflammatory response by NF-kappaB and IL-8 promotor; Upregulate chemokines such as IL-8 and RANTES; Upregulates JNK, p38 MAP kinase; Induces apoptosis and cell cycle arrest |
| ORF7b Template:UniProt |
Unknown |
| ORF8a Template:Uniprot |
Induces apoptosis through mitochondria pathway |
| ORF8b Template:UniProt |
Enhances cellular DNA synthesis, also known as X5. |
| ORF9a Template:UniProt |
Nucleocapsid (N) protein, viral RNA packaging (structural protein) |
| ORF9b Template:UniProt |
Induces apoptosis |
| ORF9c Template:UniProt |
Also known as ORF14; function unknown and may not be protein-coding |
| ORF10 Template:UniProt |
Novel gene in SARS-CoV-2, of unknown function; may not be protein-coding |
| UniProt identifiers shown for SARS-CoV proteins unless they are specific to SARS-CoV-2 | |
The functions of several of the viral proteins are known.<ref name=McBride2012>Template:Cite journal</ref> ORFs 1a and 1b encode the replicase/transcriptase polyprotein, and later ORFs 2, 4, 5, and 9a encode, respectively, the four major structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N).<ref>Template:Cite journal</ref> The later ORFs also encode for eight unique proteins (orf3a to orf9b), known as the accessory proteins, many with no known homologues. The different functions of the accessory proteins are not well understood.<ref name=McBride2012/>
SARS coronaviruses have been genetically engineered in several laboratories.<ref>Template:Cite journal</ref>
Phylogenetics

Phylogenetic analysis showed that the evolutionary branch composed of Bat coronavirus BtKY72 and BM48-31 was the base group of SARS–related CoVs evolutionary tree, which separated from other SARS–related CoVs earlier than SARS-CoV-1 and SARS-CoV-2.<ref name="pmid32007145">Template:Cite journal</ref><ref name="CVSG"/> Template:Clade
SARS-CoV-1 related
Template:SARS-CoV-1 related coronavirus
SARS-CoV-2 related
Template:SARS-CoV-2 related coronavirus
Morphology


The morphology of the SARS-related coronavirus is characteristic of the coronavirus family as a whole. The viruses are large pleomorphic spherical particles with bulbous surface projections that form a corona around the particles in electron micrographs.<ref>Template:Cite journal</ref> The size of the virus particles is in the 80–90 nm range. The envelope of the virus in electron micrographs appears as a distinct pair of electron dense shells.<ref>Template:Cite journal</ref>
The viral envelope consists of a lipid bilayer where the membrane (M), envelope (E) and spike (S) proteins are anchored.<ref name="Lai_1997">Template:Cite journal</ref> The spike proteins provide the virus with its bulbous surface projections, known as peplomers. The spike protein's interaction with its complement host cell receptor is central in determining the tissue tropism, infectivity, and species range of the virus.<ref>Template:Cite book</ref><ref>Template:Cite journal</ref>
Inside the envelope, there is the nucleocapsid, which is formed from multiple copies of the nucleocapsid (N) protein, which are bound to the positive-sense single-stranded (~30 kb) RNA genome in a continuous beads-on-a-string type conformation.<ref>Template:Cite book</ref><ref>Template:Cite journal</ref> The lipid bilayer envelope, membrane proteins, and nucleocapsid protect the virus when it is outside the host.<ref>Template:Cite journal</ref>
Life cycle
SARS-related coronavirus follows the replication strategy typical of all coronaviruses.<ref name = "Fehr_2015" /><ref>Template:Cite book</ref>
Attachment and entry

The attachment of the SARS-related coronavirus to the host cell is mediated by the spike protein and its receptor.<ref name=":2">Template:Cite book</ref> The spike protein receptor binding domain (RBD) recognizes and attaches to the angiotensin-converting enzyme 2 (ACE2) receptor.<ref name="pmid24172901"/> Following attachment, the virus can enter the host cell by two different paths. The path the virus takes depends on the host protease available to cleave and activate the receptor-attached spike protein.<ref name=":6">Template:Cite journal</ref> A notably difference between SARS-CoV-1 and SARS-CoV-2, is that SARS-CoV-2 is pre-cleaved due to its furin cleavage site.<ref>Template:Cite journal</ref>
The attachment of sarbecoviruses to ACE2 has been shown to be an evolutionarily conserved feature, present in many (but not all) species of the taxon with ACE2 using representatives in Africa, Asia, and Europe.<ref>Template:Cite journal</ref>
The first path the SARS coronavirus can take to enter the host cell is by endocytosis and uptake of the virus in an endosome. The receptor-attached spike protein is then activated by the host's pH-dependent cysteine protease cathepsin L. Activation of the receptor-attached spike protein causes a conformational change, and the subsequent fusion of the viral envelope with the endosomal wall.<ref name=":6" />
Alternatively, the virus can enter the host cell directly by proteolytic cleavage of the receptor-attached spike protein by the host's TMPRSS2 or TMPRSS11D serine proteases at the cell surface.<ref>Template:Cite journal</ref><ref>Template:Cite journal</ref> In the SARS coronavirus, the activation of the C-terminal part of the spike protein triggers the fusion of the viral envelope with the host cell membrane by inducing conformational changes which are not fully understood.<ref name="pmid316509562">Template:Cite journal</ref>
Genome translation
| Function of coronavirus nonstructural proteins (nsps)<ref name=":1">Template:Cite book</ref> | |
|---|---|
| Protein | Function |
| nsp1 | Promotes host mRNA degradation, blocks host translation;<ref>Template:Cite journal</ref> blocks innate immune response |
| nsp2 | Binds to prohibitin proteins; unknown function |
| nsp3 | Multidoman transmembrane protein; interacts with N protein; promotes cytokine expression; PLPro domain cleaves polyprotein pp1ab and blocks host's innate immune response; other domains unknown functions |
| nsp4 | Transmembrane scaffold protein; allows proper structure for double membrane vesicles (DMVs) |
| nsp5 | 3CLPro cleaves polyprotein pp1ab |
| nsp6 | Transmembrane scaffold protein; unknown function |
| nsp7 | Forms hexadecameric complex with nsp8; processivity clamp for RdRp (nsp12) |
| nsp8 | Forms hexadecameric complex with nsp7; processivity clamp for RdRp (nsp12); acts as a primase |
| nsp9 | RNA-binding protein (RBP) |
| nsp10 | nsp16 and nsp14 cofactor; forms heterodimer with both; stimulates 2-O-MT (nsp16) and ExoN (nsp14) activity |
| nsp11 | Unknown function |
| nsp12 | RNA-dependent RNA polymerase (RdRp) |
| nsp13 | RNA helicase, 5′ triphosphatase |
| nsp14 | N7 Methyltransferase, 3′-5′ exoribonuclease (ExoN); N7 MTase adds 5′ cap, ExoN proofreads genome |
| nsp15 | Endoribonuclease (NendoU) |
| nsp16 | 2′-O-Methyltransferase (2-O-MT); protects viral RNA from MDA5 |
After fusion the nucleocapsid passes into the cytoplasm, where the viral genome is released.<ref name=":2" /> The genome acts as a messenger RNA, and the cell's ribosome translates two-thirds of the genome, which corresponds to the open reading frame ORF1a and ORF1b, into two large overlapping polyproteins, pp1a and pp1ab.
The larger polyprotein pp1ab is a result of a -1 ribosomal frameshift caused by a slippery sequence (UUUAAAC) and a downstream RNA pseudoknot at the end of open reading frame ORF1a.<ref>Template:Cite journal</ref> The ribosomal frameshift allows for the continuous translation of ORF1a followed by ORF1b.<ref name=":3" />
The polyproteins contain their own proteases, PLpro and 3CLpro, which cleave the polyproteins at different specific sites. The cleavage of polyprotein pp1ab yields 16 nonstructural proteins (nsp1 to nsp16). Product proteins include various replication proteins such as RNA-dependent RNA polymerase (RdRp), RNA helicase, and exoribonuclease (ExoN).<ref name=":3">Template:Cite book</ref>
The two SARS-CoV-2 proteases (PLpro and 3CLpro) also interfere with the immune system response to the viral infection by cleaving three immune system proteins. PLpro cleaves IRF3 and 3CLpro cleaves both NLRP12 and TAB1. "Direct cleavage of IRF3 by NSP3 could explain the blunted Type-I IFN response seen during SARS-CoV-2 infections while NSP5 mediated cleavage of NLRP12 and TAB1 point to a molecular mechanism for enhanced production of IL-6 and inflammatory response observed in COVID-19 patients."<ref>Template:Cite journal</ref>
Replication and transcription
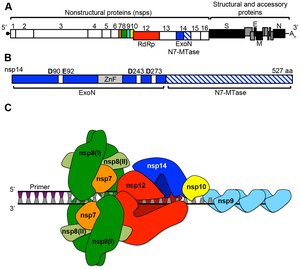
A number of the nonstructural replication proteins coalesce to form a multi-protein replicase-transcriptase complex (RTC).<ref name=":3" /> The main replicase-transcriptase protein is the RNA-dependent RNA polymerase (RdRp). It is directly involved in the replication and transcription of RNA from an RNA strand. The other nonstructural proteins in the complex assist in the replication and transcription process.<ref name=":1" />
The protein nsp14 is a 3'-5' exoribonuclease which provides extra fidelity to the replication process. The exoribonuclease provides a proofreading function to the complex which the RNA-dependent RNA polymerase lacks. Similarly, proteins nsp7 and nsp8 form a hexadecameric sliding clamp as part of the complex which greatly increases the processivity of the RNA-dependent RNA polymerase.<ref name=":1" /> The coronaviruses require the increased fidelity and processivity during RNA synthesis because of the relatively large genome size in comparison to other RNA viruses.<ref name="SextonSmith2016">Template:Cite journal</ref>
One of the main functions of the replicase-transcriptase complex is to transcribe the viral genome. RdRp directly mediates the synthesis of negative-sense subgenomic RNA molecules from the positive-sense genomic RNA. This is followed by the transcription of these negative-sense subgenomic RNA molecules to their corresponding positive-sense mRNAs.<ref name=":5">Template:Cite book</ref>
The other important function of the replicase-transcriptase complex is to replicate the viral genome. RdRp directly mediates the synthesis of negative-sense genomic RNA from the positive-sense genomic RNA. This is followed by the replication of positive-sense genomic RNA from the negative-sense genomic RNA.<ref name=":5" /> SARS-CoV replication and transcription mainly occurs in virus-induced double-membrane vesicles (DMVs) made from altered host endoplasmic reticulum.<ref name="Andronov2024">Template:Cite journal</ref>
The replicated positive-sense genomic RNA becomes the genome of the progeny viruses. The various smaller mRNAs are transcripts from the last third of the virus genome which follows the reading frames ORF1a and ORF1b. These mRNAs are translated into the four structural proteins (S, E, M, and N) that will become part of the progeny virus particles and also eight other accessory proteins (orf3 to orf9b) which assist the virus.<ref>Template:Cite book</ref>
Recombination
When two SARS-CoV genomes are present in a host cell, they may interact with each other to form recombinant genomes that can be transmitted to progeny viruses. Recombination likely occurs during genome replication when the RNA polymerase switches from one template to another (copy choice recombination).<ref name = Zhang2005/> Human SARS-CoV appears to have had a complex history of recombination between ancestral coronaviruses that were hosted in several different animal groups.<ref name="Zhang2005">Zhang XW, Yap YL, Danchin A. Testing the hypothesis of a recombinant origin of the SARS-associated coronavirus. Arch Virol. 2005 Jan;150(1):1-20. Epub 2004 Oct 11. PMID 15480857</ref><ref>Stanhope MJ, Brown JR, Amrine-Madsen H. Evidence from the evolutionary analysis of nucleotide sequences for a recombinant history of SARS-CoV. Infect Genet Evol. 2004 Mar;4(1):15-9. PMID 15019585</ref>
Assembly and release
RNA translation occurs inside the endoplasmic reticulum. The viral structural proteins S, E and M move along the secretory pathway into the Golgi intermediate compartment. There, the M proteins direct most protein-protein interactions required for assembly of viruses following its binding to the nucleocapsid.<ref name=":4">Template:Cite book</ref> Progeny viruses are released from the host cell by exocytosis through secretory vesicles.<ref name=":4" />
See also
- Bat SARS-like coronavirus WIV1 (SL-CoV-WIV1)
- Bat SARS-like coronavirus RsSHC014
- Bat coronavirus RaTG13
- Civet SARS-CoV
Notes
References
Further reading
- Template:Cite journal
- Template:Cite journal
- Template:Cite journal
- Template:Cite journal
- Template:Cite journal
- Template:Cite book
- Template:Cite book
External links
- Template:Commons category-inline
- Template:Wikispecies-inline
- WHO press release identifying and naming the SARS virus (archived 23 April 2003)
- The SARS virus genetic map Template:Webarchive
- Science special on the SARS virus (free content: no registration required)
- Template:Webarchive
- U.S. Centers for Disease Control and Prevention (CDC) SARS home (archived 12 April 2016)
Template:Coronaviridae Template:SARS Template:Viral diseases Template:Zoonotic viral diseases Template:Taxonbar Template:Authority control